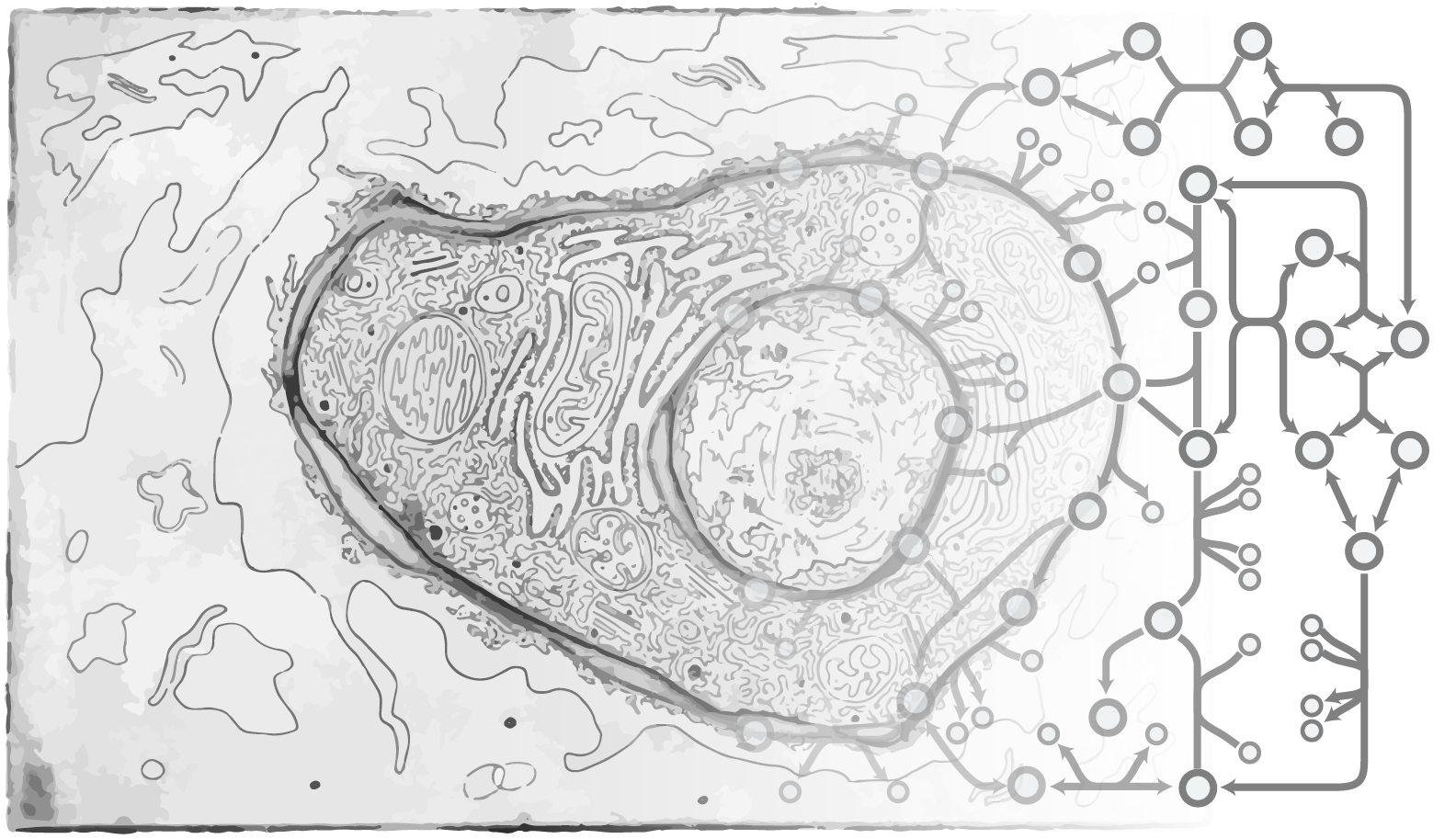
In the mammalian genome, roughly 1/3 of the protein-coding genes make products that directly mediate most interactions between cells, organs, and pathogens. These include hormones, membrane proteins, immune factors, extracellular matrix proteins, enzymes, etc. All of these proteins are synthesized and secreted through the secretory pathway, a system localized predominantly to the endoplasmic reticulum, Golgi apparatus, and other components of the endomembrane system in eukaryotic cells. Hundreds of proteins are known to facilitate protein production in the secretory pathway, but the complexity of the pathway obfuscates how each part of the pathway impacts the synthesis and secretion of the various hormones, antibodies, and diverse proteins. Indeed, it is expected that only subsets of the secretory pathway machinery will be necessary to synthesize each protein. Can we capture all of the steps and unique machinery required to make any given secreted protein? A detailed systems-level view of the secretory pathway would be invaluable to unravel how protein synthesis is controlled and to help elucidate the causes of amyloid diseases, to control post-translational modifications such as glycosylation, and to aid in engineering mammalian cells that more efficiently produce immunotherapies and other high-value biopharmaceuticals.
To begin addressing these questions, we are establishing a systems biology platform to study the mammalian secretory pathway. This includes the generation of experimental omics data and the development of systems biology models of the pathway (Gutierrez and Lewis, Biotech J, 2015; Kuo, Curr Opin in Biotech). As one example, we have developed a systems-level mechanistic understanding of the mammalian secretory pathway is necessary given its importance in the synthesis, folding and delivery of recombinant proteins in Chinese hamster ovary (CHO) cells. With this model, we can estimate the theoretical energetic trade-off between growth and productivity in CHO cells producing a set of recombinant proteins with relevance in the biopharmaceutical industry. We have shown a dependence of the growth-productivity trade-off upon protein size, amino acid composition, and complexity. This model can be used to predict potential knock-out targets that could free up resources and boost productivity.