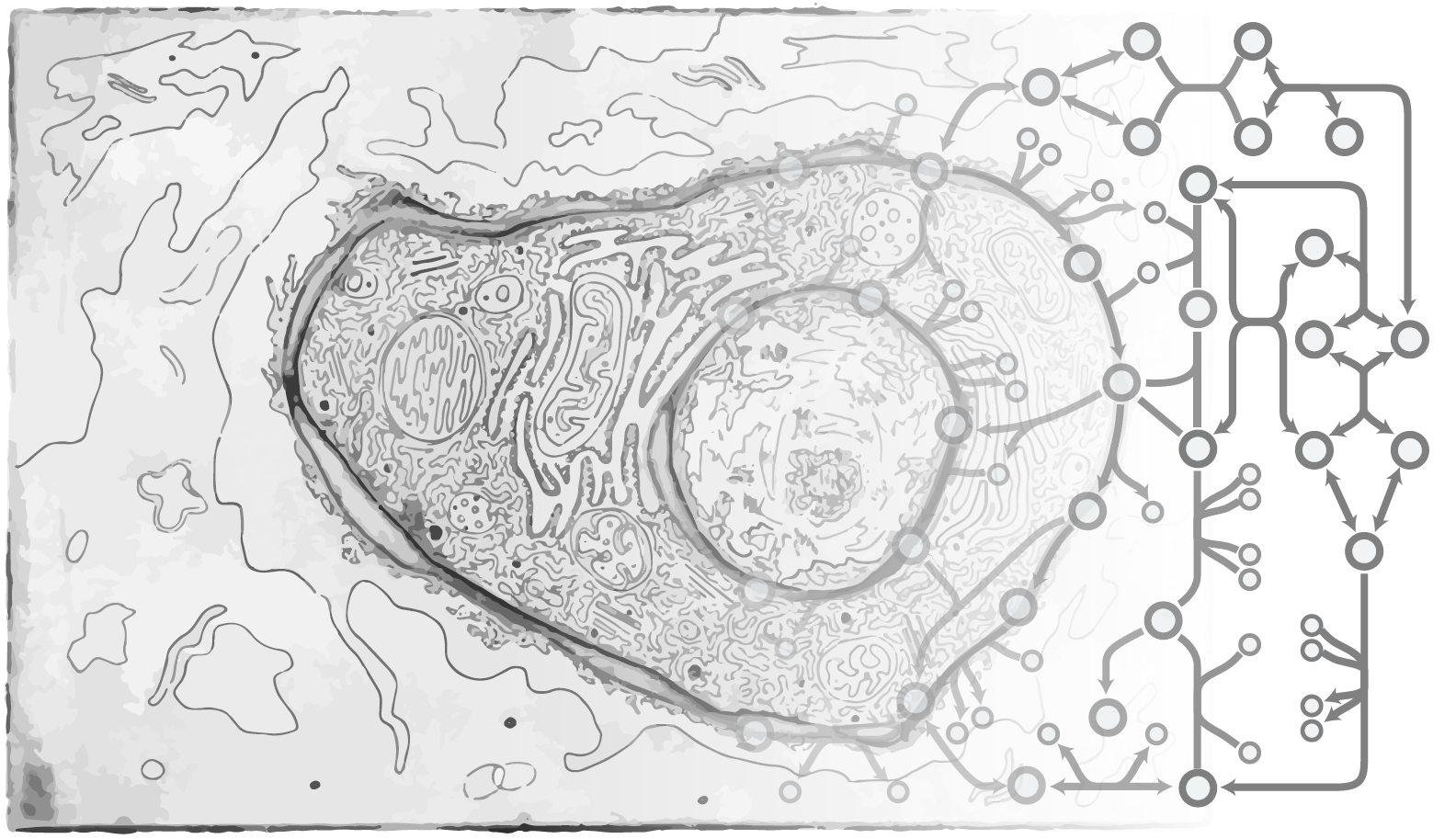
Protein Secretion Pathway in Mammalian Cells
 Identification of protein–protein interactions (PPIs) is a fundamental approach to study molecular and biological mechanisms within a cell. The secretory pathway in mammalian cells is highly complex and involves many PPIs that assure proper folding and post-translational modifications. However, expression of recombinant proteins with different physicochemical properties have diverse impact on secretory pathway which usually results in cellular stress that impairs the protein quality and quantity. My research centers on using BioID technology to investigate PPIs that regulates the function of the secretory pathway in mammalian cells during expressing diverse recombinant proteins. This enable us to understand how cells tune the interactome in the secretory pathway to dynamically cope with the cellular stress and ensure efficient protein expression and maintain cellular homeostasis. These findings can be used to rationally engineer mammalian cells to enhance quality and expression of difficult-to-express proteins.
Identification of protein–protein interactions (PPIs) is a fundamental approach to study molecular and biological mechanisms within a cell. The secretory pathway in mammalian cells is highly complex and involves many PPIs that assure proper folding and post-translational modifications. However, expression of recombinant proteins with different physicochemical properties have diverse impact on secretory pathway which usually results in cellular stress that impairs the protein quality and quantity. My research centers on using BioID technology to investigate PPIs that regulates the function of the secretory pathway in mammalian cells during expressing diverse recombinant proteins. This enable us to understand how cells tune the interactome in the secretory pathway to dynamically cope with the cellular stress and ensure efficient protein expression and maintain cellular homeostasis. These findings can be used to rationally engineer mammalian cells to enhance quality and expression of difficult-to-express proteins.
CHO cell genomics and Engineering
Despite the improvements in protein production in CHO cells over the past decades, quantities of difficult-to-express-proteins as well as protein quality still require to be enhanced, particularly through cell-based engineering strategies. However, accurate and well-annotated genomic resources is essentials to facilitate CHO genome based cell engineering efforts. My research focuses on enhancing genome annotation by identification of Transcription Start Site (TSS) in CHO cells genome using innovative sequencing tools. TSS identification enables us to design CRISPR-based activation and repression libraries to study the functional role of genes involved in protein secretion and post translation modifications in CHO cells.
Link to my Google Scholar
Contact : msamoudi (at) ucsd (dot) edu