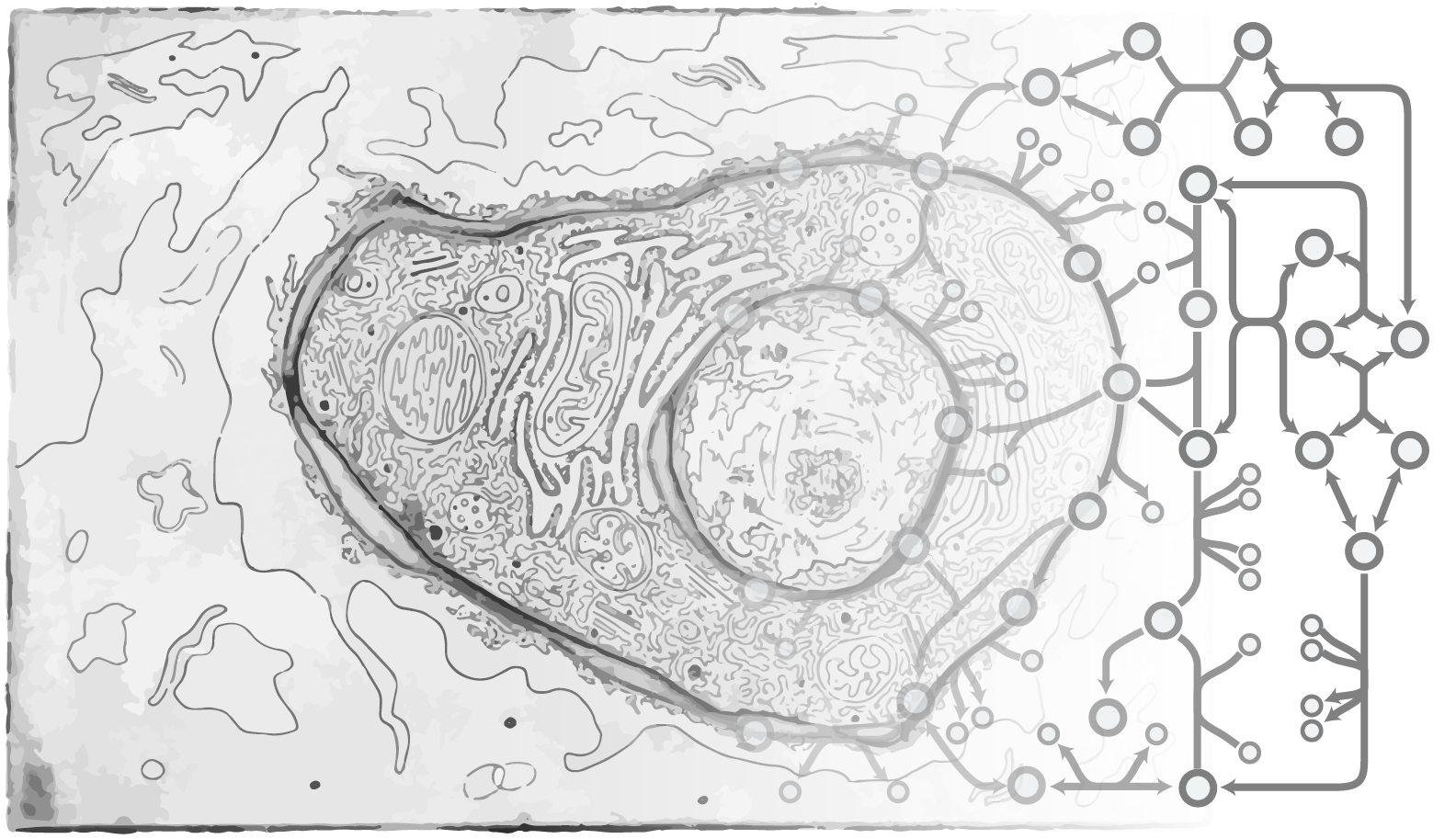
My current research focuses on genomic instability, with special applications in engineering of CHO cells, the primary cell line used in pharmaceutical production. I use various methods like cell culture, CRISPR/Cas9 genome editing, microscopy, sequencing, and bioinformatics.
CURRENT PROJECTS:
Improving genomic stability in CHO cells
CHO cells are the primary production host of therapeutic proteins. However, their inherent genomic instability poses a problem for protein manufacturing since it negatively affects culture viability, productivity and product quality. The primary cause of genomic instability is deficient DNA repair, in particular repair of double-strand breaks. This project aims at quantification of DNA repair capabilities in various CHO cell lines, and the identification of targets whose genetic engineering has a potential to enhance DNA repair, improve genomic stability, and thereby greatly facilitate industrial protein manufacturing.
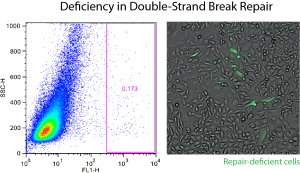
Automated analysis of sequencing data from CRISPR/Cas9 screens
CRISPR/Cas9 screens have become an extremely popular tool for biomedical discovery, but processing and analysis of the sequencing data produced by these screens pose a significant challenge for researchers lacking computational support. In this project, we developed PinAPL-Py, an online workflow for the analysis of such screens that can be operated through easy drag-and-drop and run without bioinformatic expertise.
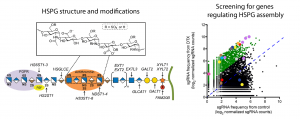
Unraveling the genetic control of Heparan-sulfate proteoglycans using genome-wide screens
Heparan-sulfate proteoglycans (HSPGs) are major components of the extracellular matrix and involved in a wide variety of cellular functions, e.g. as co-receptors for EGF-related signaling pathways. Although its biosynthesis is fairly well understood, very little is known about its genetic control and tissue-specific expression. In collaboration with the Esko laboratory, we utilize genome-wide CRISPR/Cas9 knock-out screens to find genes controlling HSPG assembly and modification.
Glycoengineering in CHO cells
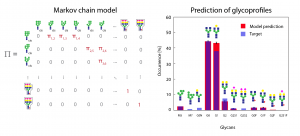
Glycosylation of therapeutic proteins is a critical quality attribute in their manufacturing due to its role in protein folding, stability, and efficacy. Since glycosylation does not follow a direct template, but instead is the result of a stochastic reaction network in the Golgi, its tailoring and experimental manipulation (“glycoengineering”) is a primary challenge in industrial production. This project aims at the development and application of stochastic glycosylation models to pave the way for a more rational and efficient approach to glycoengineering.
contact: pspahn at eng dot ucsd dot edu